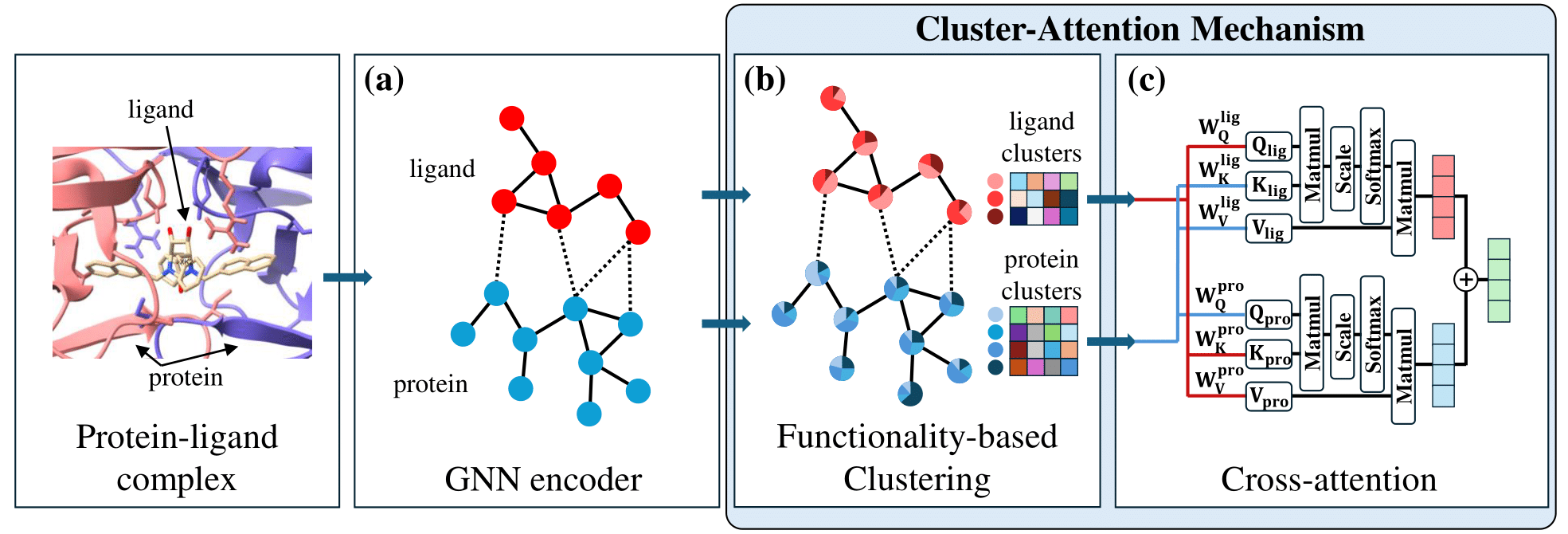
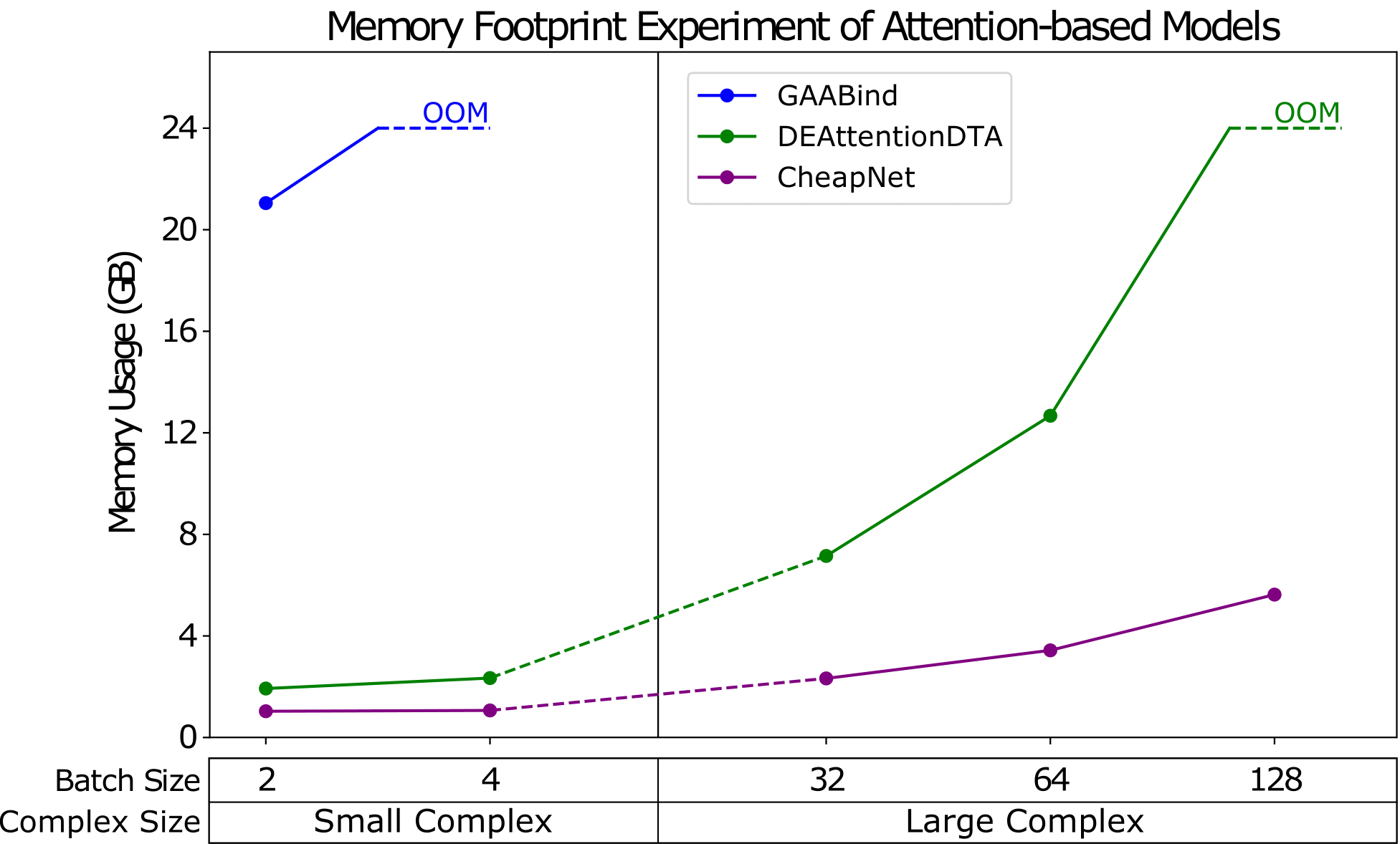
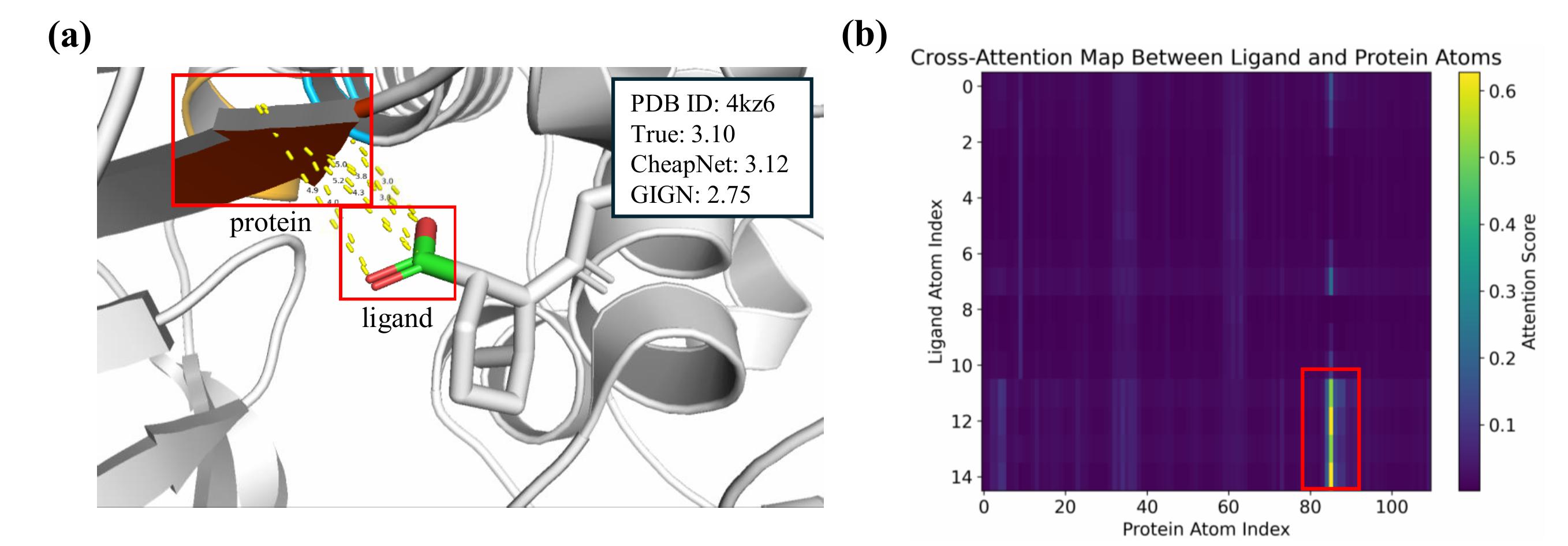
Problem Domain:
- Accurate protein-ligand binding affinity prediction for drug discovery
Existing Challenges:
- Traditional lab methods are slow and costly
- Existing models often rely solely on atom-level interactions, which suffer from:
- Noise from irrelevant atoms & Computational inefficiency
- Difficulty in determining meaningful atom clusters that drive interactions
- Previous cluster-level approaches relied on predefined clusters
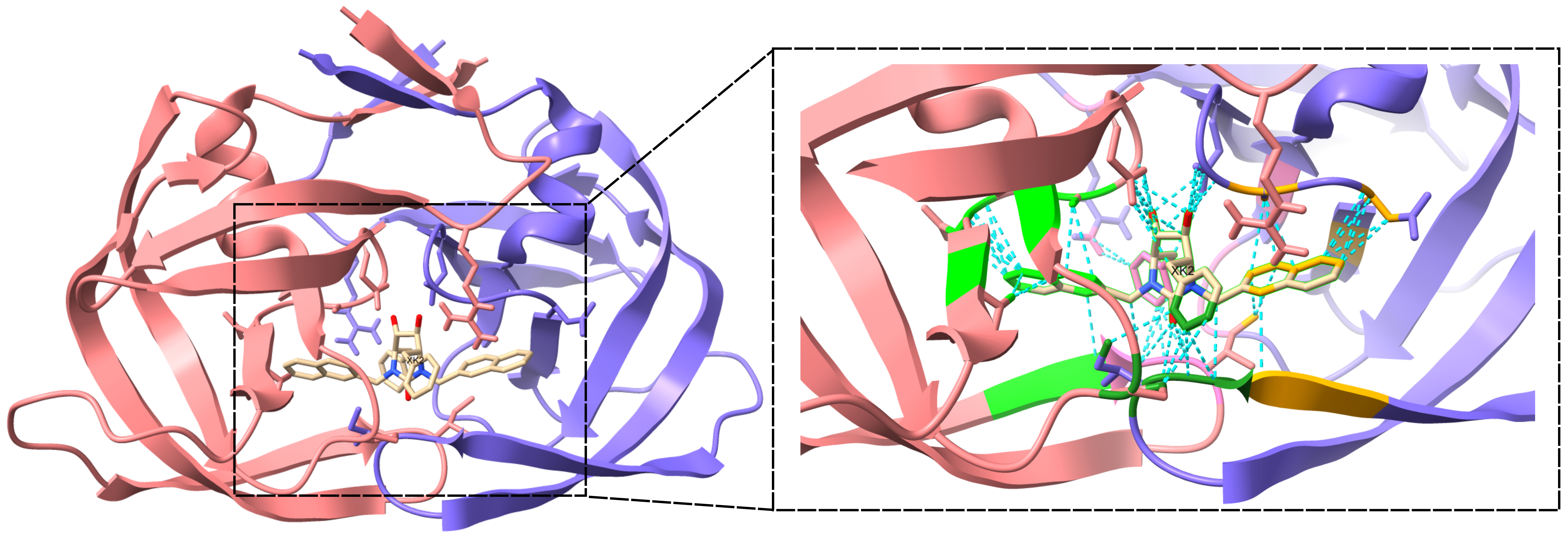
Figure 1: Illustration of protein-ligand complex and interaction patterns