
State-of-the-art equivariant Graph Neural Networks (GNNs) have significantly advanced molecular simulation by approaching quantum mechanical accuracy in predicting energies and forces. However, their substantial computational cost limits adoption in large-scale molecular dynamics simulations. Knowledge distillation (KD) offers a promising solution, but existing methods for Machine Learning Force Fields (MLFFs) often resort to simplistic atom-wise feature matching or complex second-order information distillation, overlooking fundamental first-order relational knowledge: how the teacher represents the potential energy surface (PES) through learned interatomic interactions. This paper introduces FORK, First-Order Relational Knowledge Distillation, a novel KD framework that directly distills interatomic relational knowledge by modeling each interaction as a relational vector derived from bonded atom embeddings. FORK employs contrastive learning to train students to generate relational vectors uniquely identifiable with teacher counterparts, effectively teaching the geometry of the teacher's learned PES. On the challenging OC20 benchmark, FORK enables a compact 22M-parameter student model to achieve superior energy and force prediction accuracy, significantly outperforming strong distillation baselines and demonstrating more effective transfer of physical knowledge.
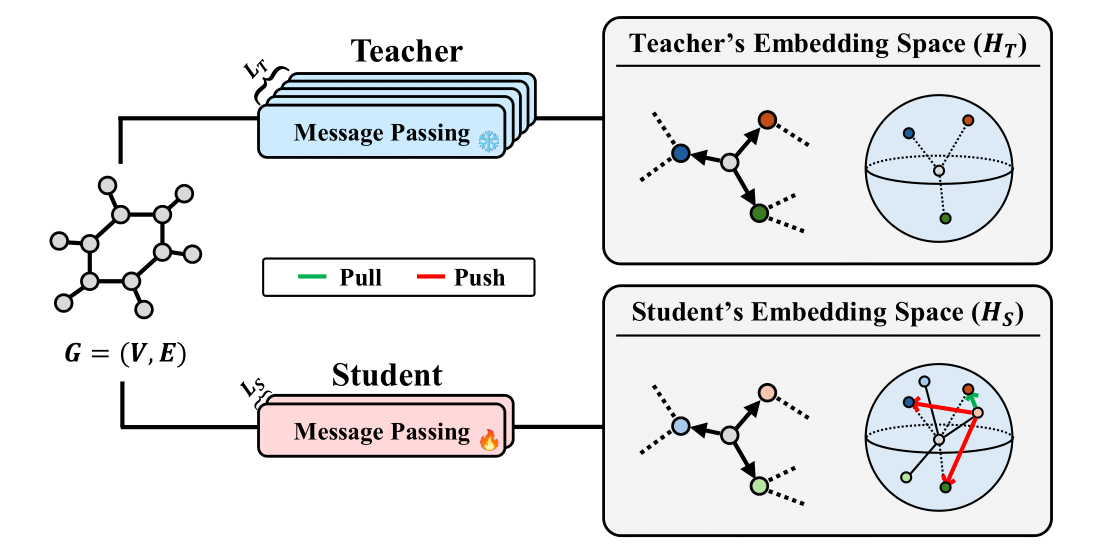
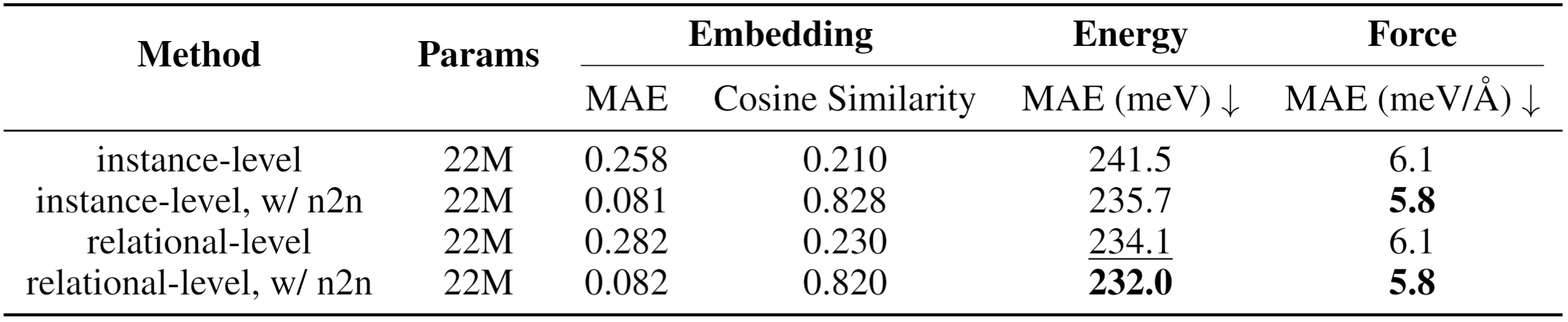
Relational Vectors: FORK models each interatomic interaction as a relational vector derived from bonded atom embeddings (e.g., zsrc - zdst). These vectors serve as proxies for the teacher's learned representation of potential along specific interactions.
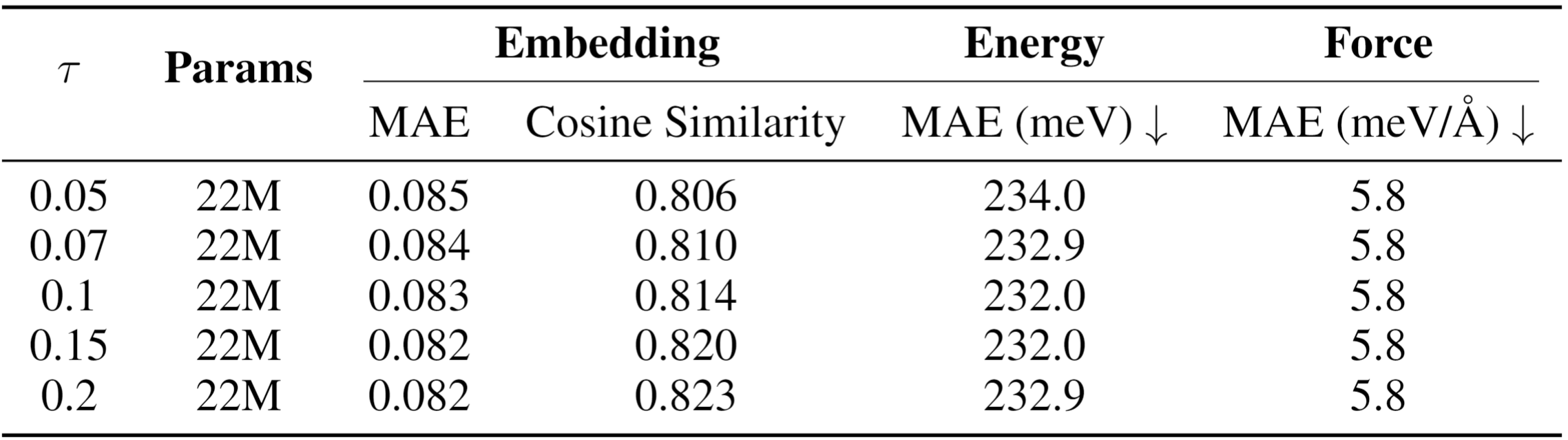
Contrastive Objective: An InfoNCE-style loss trains the student to produce relational vectors that are discriminatively similar to the teacher's corresponding vectors, effectively teaching the student the geometry of these interactions.
Physics-Informed Distillation: Unlike conventional atom-wise feature matching, FORK directly distills the fundamental physics of interatomic potentials, focusing on how atoms interact rather than treating them as isolated entities.
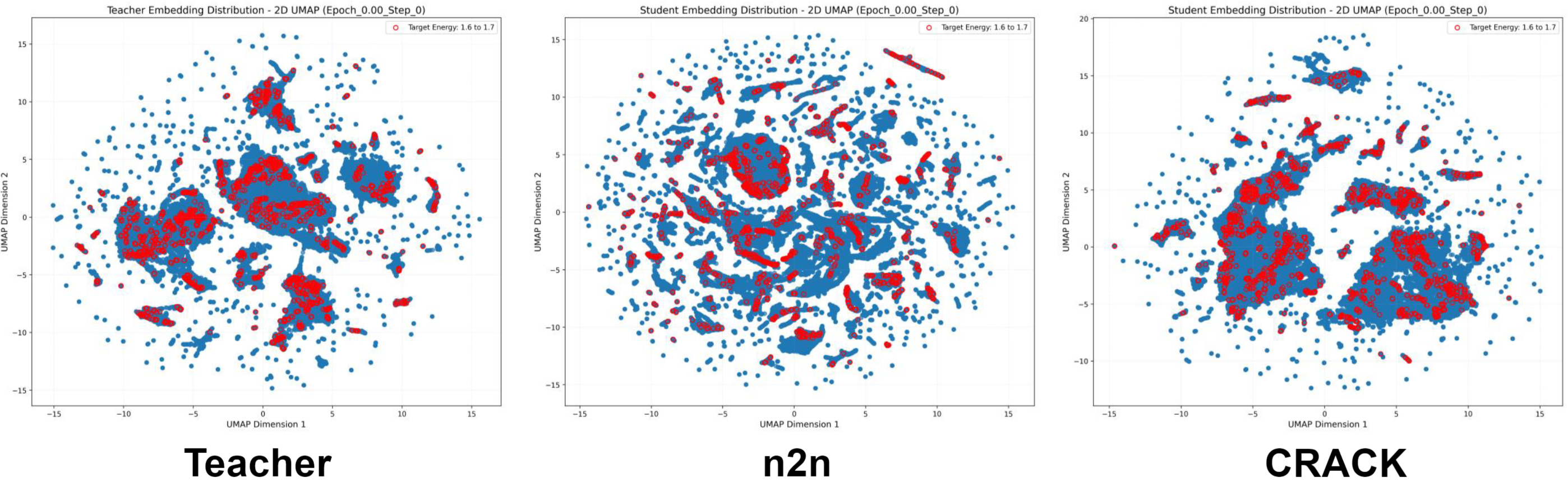
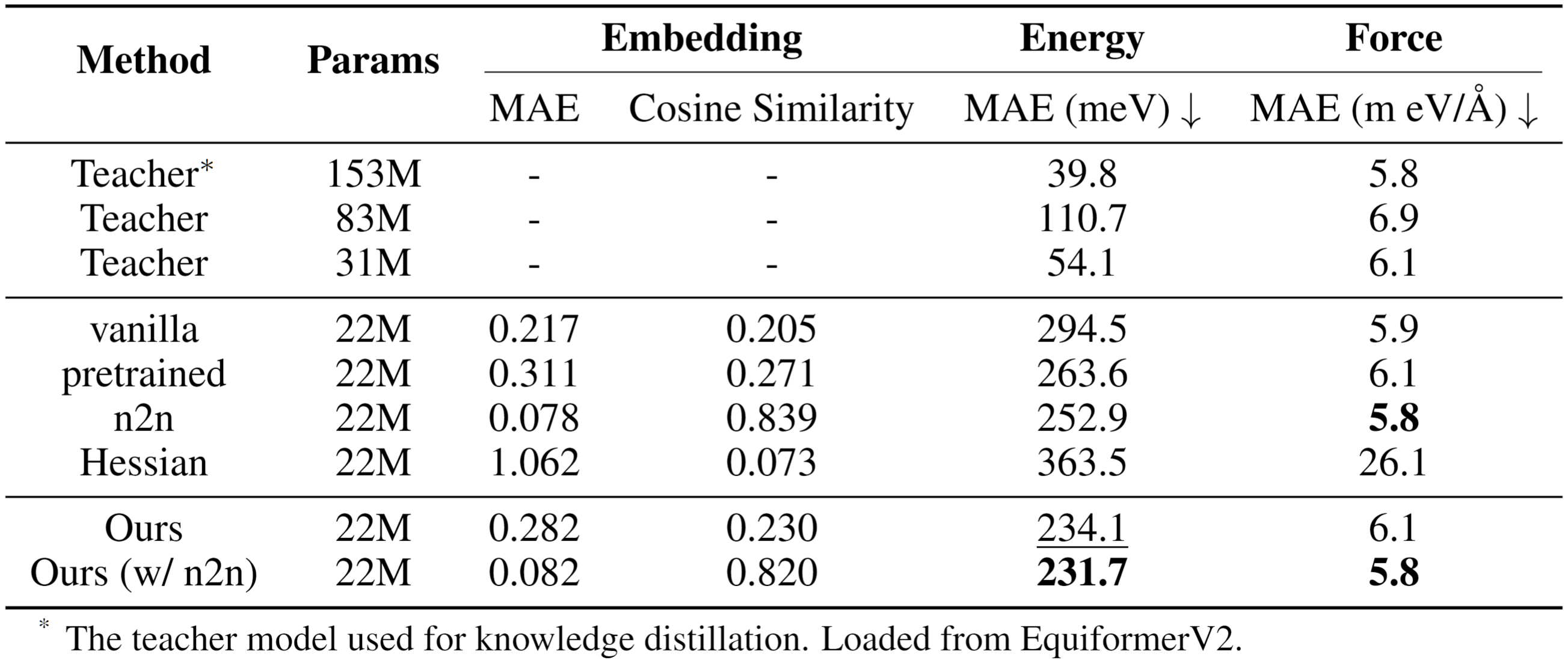
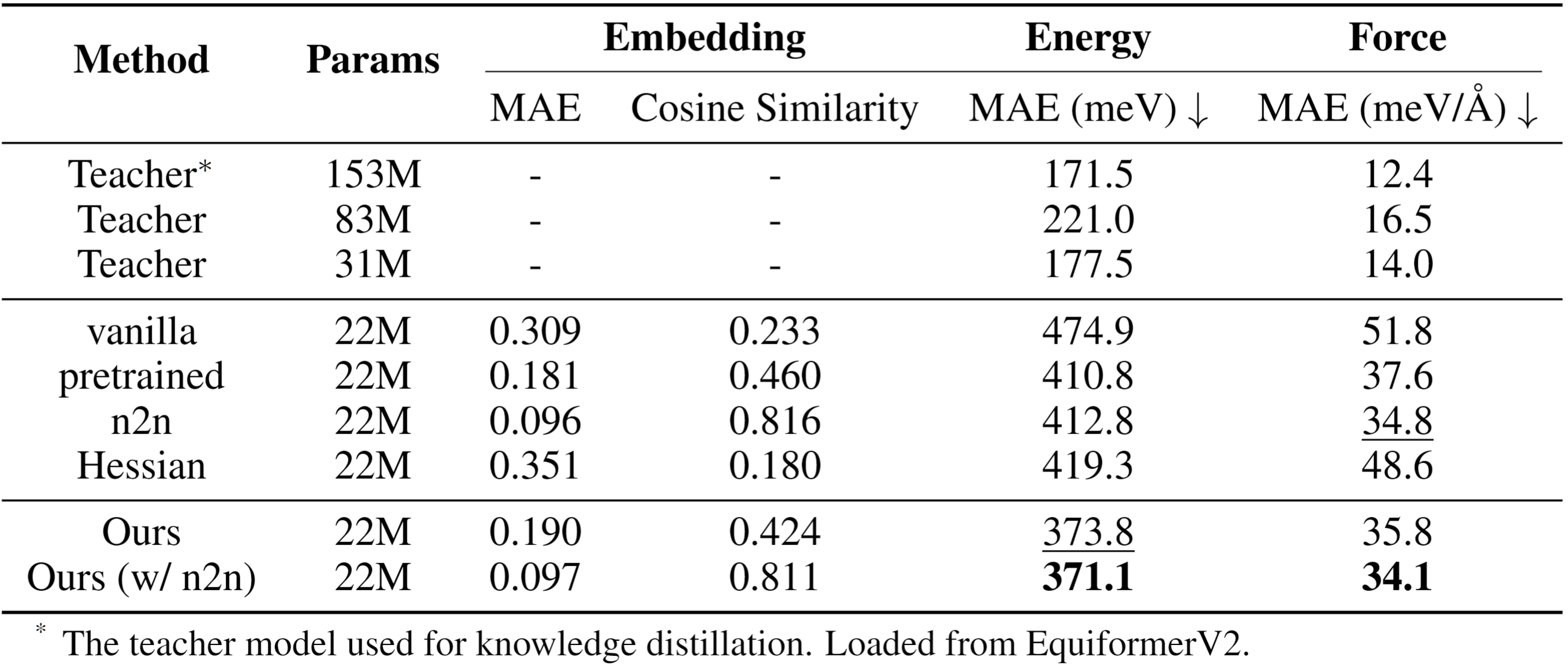
FORK achieves superior performance in energy and force prediction on the challenging OC20 benchmark. The method enables a compact 22M-parameter student model to significantly outperform strong distillation baselines, including conventional node-to-node feature matching and Hessian-based distillation. FORK demonstrates more effective transfer of physical knowledge by directly modeling the geometry of interatomic interactions learned by the teacher model.
@inproceedings{
lim2025fork,
title={FORK: First-Order Relational Knowledge Distillation for Machine Learning Interatomic Potentials},
author={Hyukjun Lim and Seokhyun Choung and Jeong Woo Han},
booktitle={arXiv},
year={2025},
}